Cinder app for iPhone and iPad
Protein crystals are an essential part of X-ray crystallography, the predominant method used to figure out the molecular structure of biologically important molecules like proteins. CSIRO (like many other research institutions) is interested in the molecular structure of proteins because it is essential for our biotechnology research.
Finding protein crystals takes a lot of time!
In order to grow a protein crystal, CSIRO must set up thousands of crystallisation experiments. These experiments are small droplets where different mixtures of chemicals are combined with a concentrated, pure protein sample. Unfortunately, there is no way of knowing which combination of chemicals will work for any particular protein, so for every protein, many different droplets are set up. Then we take photos of the experiments over many weeks, and look at them to see if crystals have formed. Less than 1% of the experiments have crystals. CSIRO developed Cinder so we can crowd source our search for experiments with crystals. We are using this information to help us develop machine image analysis tools to help us find crystals automatically. (A similar approach was used by Facebook to enable it to recognise people’s faces in pictures).
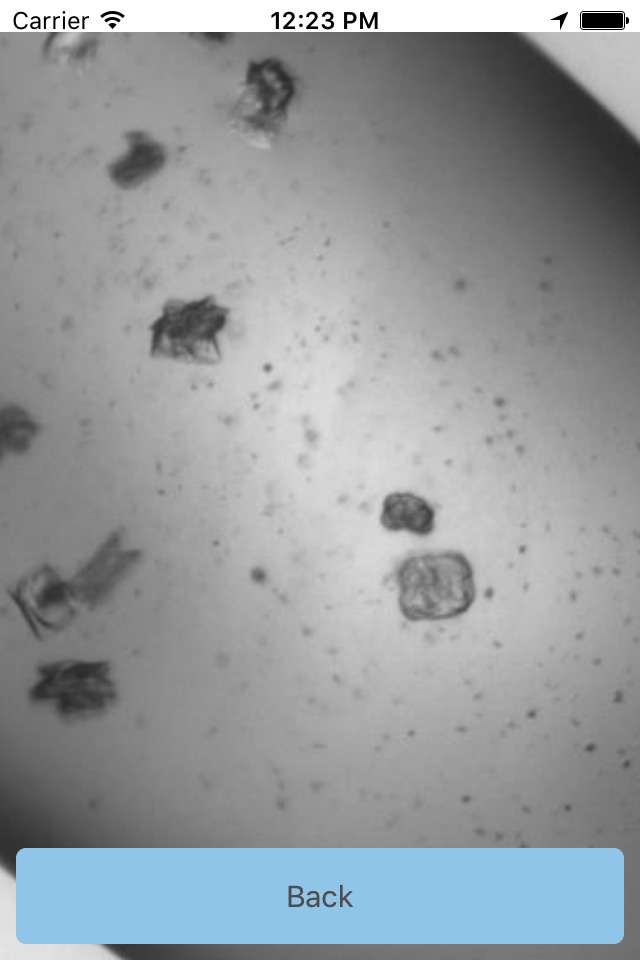
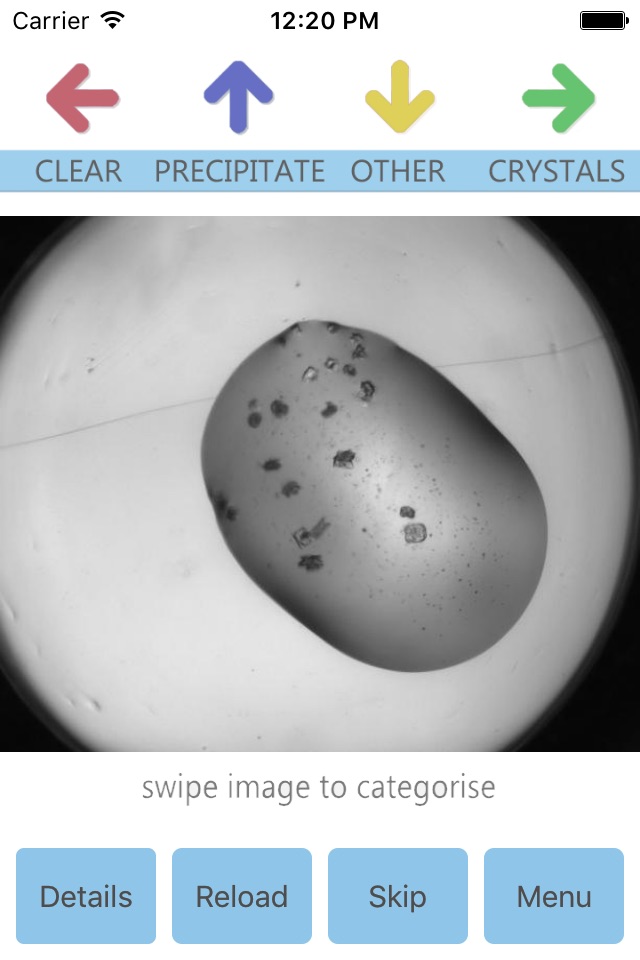
One of the challenges is that crystal can grow in many different ways, and in many different shapes. When we think of crystals we think of something like a grain of salt, or of a diamond. But crystals can also be long and thin (like spaghetti) or clusters of tiny needles – like a sea urchin. The crystals may be big and really easy to spot, or they may be hidden in a cloud of precipitated protein (almost like a fog). Thus our computer program used to find crystals can’t just look for one type of crystal, but has to be trained on a large set of different images, some with crystals, some with no crystals. Humans are very good at finding crystals in images, and we have developed Cinder so that you can help us create the training set which will be used to help the computer program learn how to find crystals.
Cinder is an easy app to use, swipe right for crystals. Your classification of each experiment is pushed back to CSIRO.
The crystals we find will be used to generate atomic structures of proteins, which are used to help scientists understand the protein. Atomic structures generated through X-ray crystallography have been used to understand how DNA works, to develop drugs against diseases like HIV, and to reveal the molecular basis of photosynthesis.
Every swipe from you is help us develop a better understanding of biology!